Uploading Metadata into Cafe Variome
There are two different ways to upload metadata into Cafe Variome.
Using the editing interface
The most straight forward way is to use the editing interface to (manually) create a new meta source. This is the recommended approach when there are only several records to add.
Using the file uploader
If there is a larger quantity of metadata, or if the metadata is being exported from another system, it's better to use the file uploader to upload the metadata. It does, however, require the metadata to be in a specific format to allow direct ingestion.
Metadata file format
The metadata file to be uploaded should be in JSON format. It should be an array containing one or multiple objects, with each object being a meta source. The structure of the meta-source should follow the meta-source model, but some fields can be omitted or left empty. The following is an example of a metadata file:
Fields and constraints
Some of the fields within the metadata model have constraints and/or formatting requirements. The following is a list of all of the fields, what they are, and their constraints. Fields not marked as optional are required, but may be left empty (as an empty string) or 0 (as a number) unless otherwise specified. All field names are case-sensitive.
Metadata entries cannot contain additional fields, except those allowed in customFields list or fields labeled as "custom." All other additional fields will be ignored and not stored or represented in any way.
Internal fields and manual assignment
The fields explained in the sections below are the fields that form the metadata about a given resource. However, there are several other fields used internally within Cafe Variome to enable specific features, like interlinking of metadata entries. These are not visible when using the editing interface, but may be manually assigned providing the data is accurate, and the admin conforms to the required processes to format them correctly.
- sourceId
The UUID of the source. If omitted, Cafe Variome will assign a UUID to the source. If present, it should be a valid UUID4 string. This is used within each Cafe Variome instance to identify the source (UUID may not be unique accross instances). It can be used to link one metadata entry to another, for example, by filling the
datasetIdsfields in the cohort model.- connectionId
The UUID of a data source this metadata entry describes. This is usually only valid when the metadata describes a dataset, but in rare cases can be assigned to other metadata. This field is not recommended to assign directly, as there is no other way to know the UUID of a data source except from checking the database.
Common fields
These are the fields that are present in all types of meta-sources.
- sourceName
stringCannot be empty.- sourceType
stringCannot be empty. Constrained vocabulary values:custom
cohort
catalog
biobank
registry
guideline
dataset
dataCollection
- resourceUrls
array[string]optionalFull URI format with schema.- publisher
objectNested JSON object.- publisherType
stringCannot be empty. Constrained vocabulary values:individual
organization
agency
other
- name
stringCannot be empty.- contactEmail
stringCannot be empty.- contactName
stringoptional.- url
stringoptionalFull URI format with schema.- location
stringoptional.
- description
stringoptionalCan be empty but not recommended.- themes
array[string]optionalURI format.- releaseLicense
stringoptionalFull URI format with schema.- language
stringoptionalTwo-character code adhering to ISO639-1 standard, lower case.- customFields
objectoptionalKey-value or key[values] pairs. Constraints:Key must be a string and cannot contain special characters:
.,$,/, or\.If a key is present, the value cannot be `null` but can be an empty string or an empty array.
Cohort specific fields
These are the fields specific to the cohort type.
- cohortDetails
objectNested JSON object.- siteType
stringCannot be empty. Constrained vocabulary values:singleSite
multiSite
multiCountry
- country
stringCannot be empty. Two-character code adhering to ISO3166-1 standard, upper case.- yearStart
integerCannot be empty. Four-digit integer.
- collectedTypes
objectoptionalNested JSON object.- participants
objectoptionalNested JSON object.- diseases
array[string]Cannot be empty. Constrained vocabulary values:controlGroup
ad
pd
irbd
dlb
caa
ftd
als
psp
cbd
msa
hd
ataxia
other
Note: An empty array will cause the entire
participantsobject to be ignored.- numberOfSubjects
integerMust be greater than 0. If 0, the entireparticipantsobject will be ignored.
- bioSamples
array[string]optionalConstrained vocabulary values:csf
serum
plasma
dna
saliva
urine
stool
- images
array[string]optionalConstrained vocabulary values:mri
petAmyloid
petTau
spect
ocular
- cognitiveData
array[string]optionalConstrained vocabulary values:crossSectional
longitudinal
- datasetIds
array[string]optionalUUID format. Datasets must be either present in the same file or already uploaded to the system.
Dataset Specific fields
These are the fields specific to the dataset type.
- datasetVersions
array[object]Nested array of JSON objects.- datasetDetails
objectNested JSON object.- versionId
stringoptionalValid UUID 4 string. If omitted, a UUID will be automatically assigned.- versionName
stringoptionalSemantic versioning recommended. Non-semantic versions will disable version parsing, comparison, and sorting.- keywords
array[string]optionalArray of keyword strings.- publishedDate
stringoptionalDate string in the format of `YYYY-MM-DD`.- updateDate
stringoptionalDate string in the format of `YYYY-MM-DD`.
- datasetContent
objectNested JSON object.- numberOfSubjects
integerMust be greater than 0. May be an approximate number if exact count is confidential.- minAge
integeroptionalMust be greater than 0.- maxAge
integeroptionalMust be greater than 0.- countries
array[string]optionalArray of two-character codes adhering to ISO3166-1 standard, upper case.- diseases
array[string]optionalConstrained vocabulary values:controlGroup
ad
pd
irbd
dlb
caa
ftd
als
psp
cbd
msa
hd
ataxia
other
- sex
array[string]optionalConstrained vocabulary values:male
female
other
undifferential
unknown
- clinical
array[string]optionalConstrained vocabulary values:comorbidities
medicationUse
familyHistory
ageOfSymptomOnset
clinicalDiagnosis
exposure
lifeStyleInfo
vitalSigns
- markers
array[string]optionalConstrained vocabulary values:amyloid
tau
neurofilamentLightChain
alphaSynuclein
dat
- images
array[string]optionalConstrained vocabulary values:mri
petAmyloid
petTau
spect
ocular
- electrophysiology
array[string]optionalConstrained vocabulary values:eeg
meg
erp
- dataTypes
array[string]optionalConstrained vocabulary values:demographics
clinical
lifestyle
functionalRatings
motor
neuropsychiatric
neuropsychological
qualityOfLife
sleepScales
digitalData
imaging
electrophysiology
neuroPathology
other
Data collection Specific fields
These are the fields specific to the data collection type.
- dataCollectionDetails
objectoptionalNested JSON object.- keywords
array[string]optionalArray of keyword strings.- publishedDate
stringoptionalDate string in the format of `YYYY-MM-DD`.- updateDate
stringoptionalDate string in the format of `YYYY-MM-DD`.
- dataCollectionContent
objectoptionalNested JSON object.- numberOfSubjects
integerMust be greater than 0. May be an approximate number if exact count is confidential.- minAge
integeroptionalMust be greater than 0.- maxAge
integeroptionalMust be greater than 0.- countries
array[string]optionalArray of two-character codes adhering to ISO3166-1 standard, upper case.- diseases
array[string]optionalConstrained vocabulary values:controlGroup
ad
pd
irbd
dlb
caa
ftd
als
psp
cbd
msa
hd
ataxia
other
- sex
array[string]optionalConstrained vocabulary values:male
female
other
undifferential
unknown
- clinical
array[string]optionalConstrained vocabulary values:comorbidities
medicationUse
familyHistory
ageOfSymptomOnset
clinicalDiagnosis
exposure
lifeStyleInfo
vitalSigns
- markers
array[string]optionalConstrained vocabulary values:amyloid
tau
neurofilamentLightChain
alphaSynuclein
dat
- images
array[string]optionalConstrained vocabulary values:mri
petAmyloid
petTau
spect
ocular
- electrophysiology
array[string]optionalConstrained vocabulary values:eeg
meg
erp
- dataTypes
array[string]optionalConstrained vocabulary values:demographics
clinical
lifestyle
functionalRatings
motor
neuropsychiatric
neuropsychological
qualityOfLife
sleepScales
digitalData
imaging
electrophysiology
neuroPathology
other
Metadata model examples
Here are some minimum and maximum examples of metadata models for different source types.
Minimum example for custom type:
Maximum example for custom type:
Minimum example for cohort type:
Maximum example for cohort type:
Minimum example for dataset type:
Maximum example for dataset type:
Minimum example for dataCollection type:
Maximum example for dataCollection type:
Uploading the metadata file
To upload the metadata file, go to the admin interface, and click on the "Create Meta Source" button.
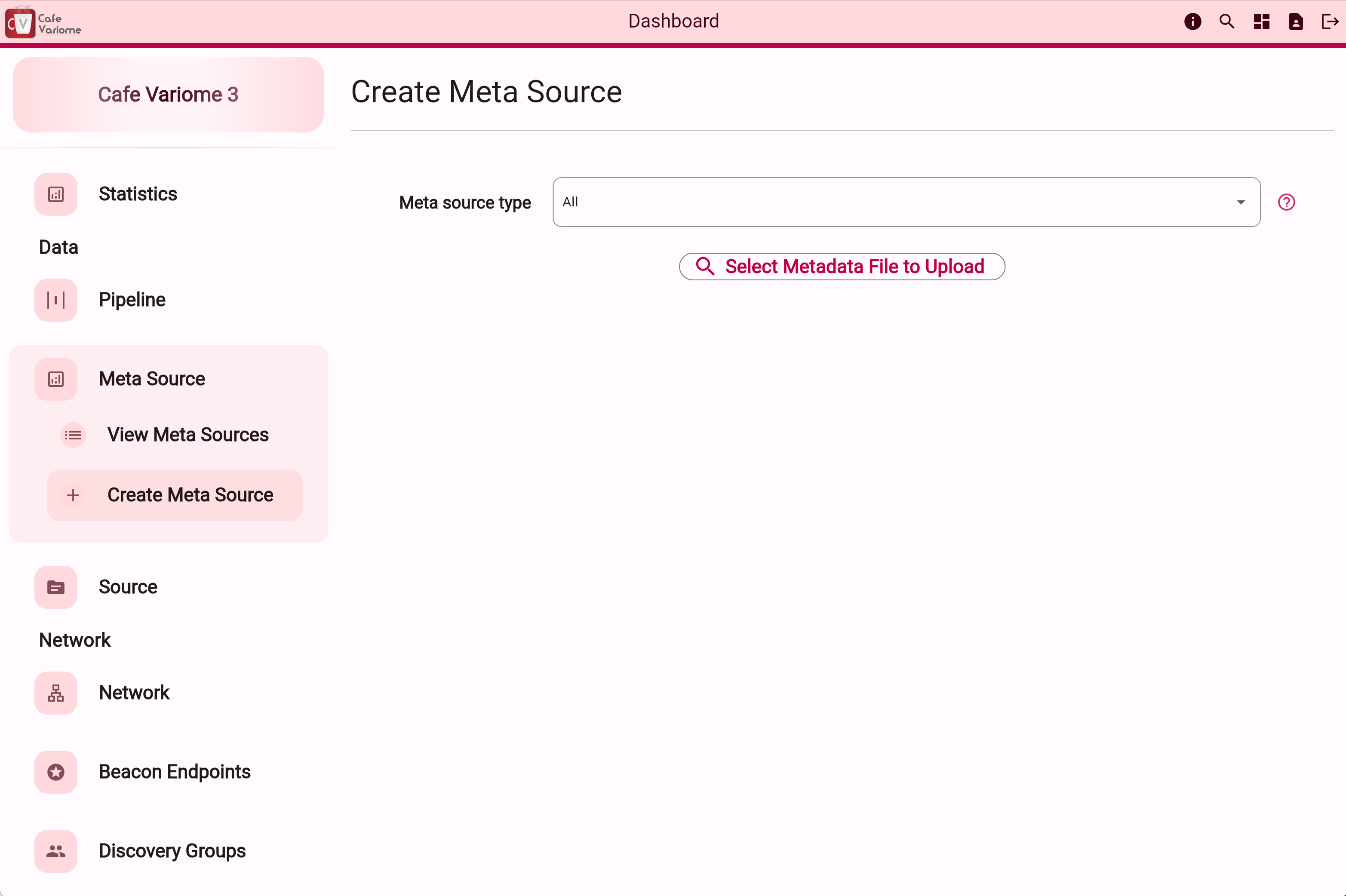
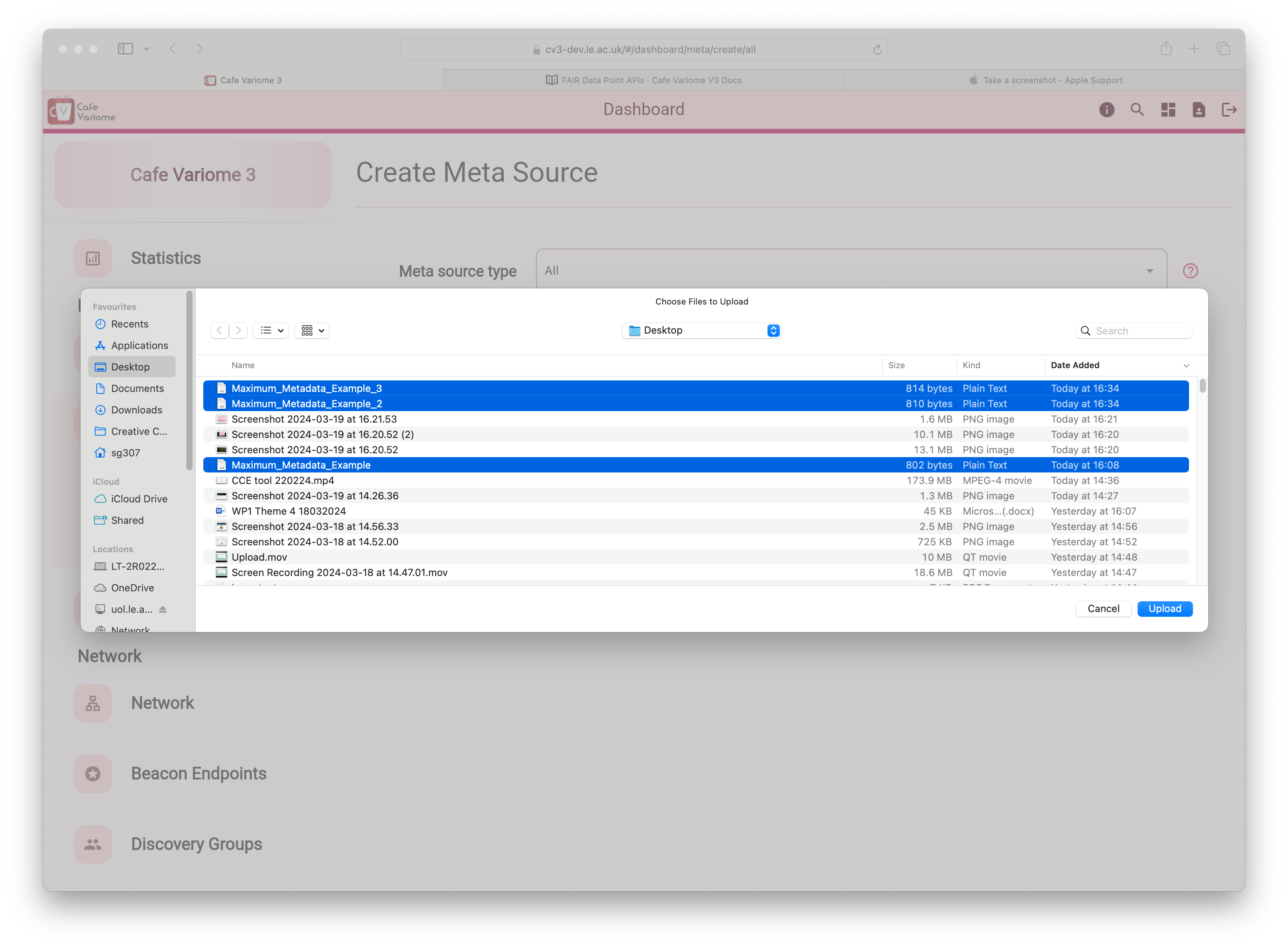
Then, set the meta source type to "all" to use the file uploader. Multiple files can be selected, and selected files can be removed before finalising the upload.
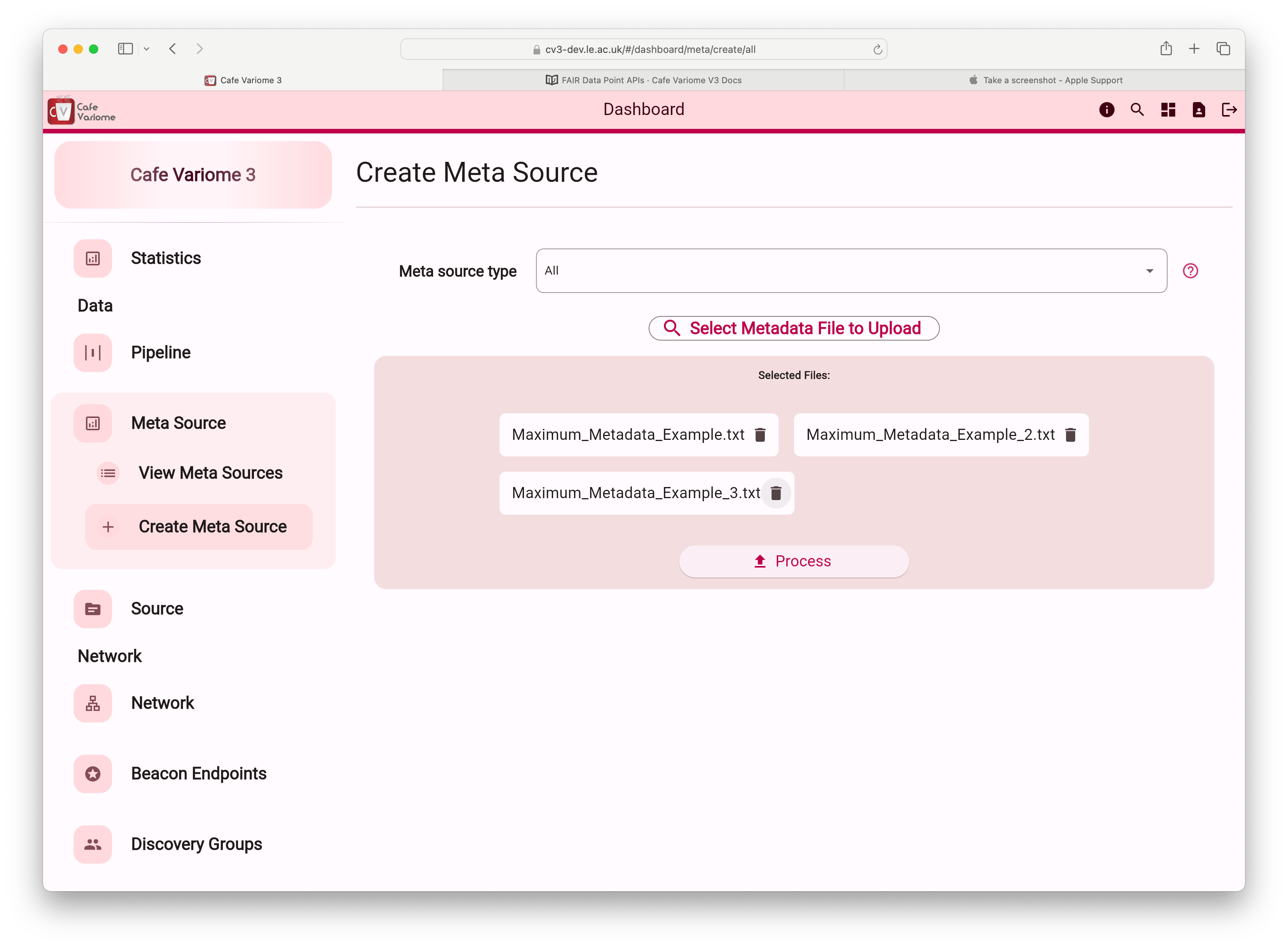
After selecting the files, click the "Process" button to begin reading and processing them. All metadata entries will first be read by the frontend, sanitized, and validated, then sent to the server for storage. Once processing is complete, you'll see a prompt displaying the number of metadata entries read and asking you to confirm the upload. After your confirmation, the metadata entries will be stored in the database and become available for search and discovery.